Research
Understanding the sequence code underlying regulatory regions in glucose regulation disorders
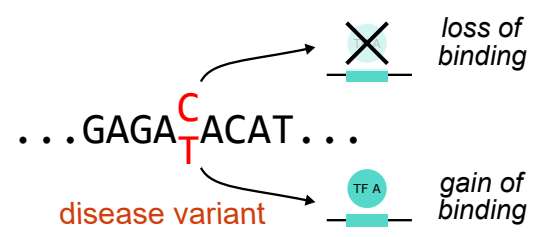
We have recently been awarded a Wellcome Career Development Award to study the sequence code underlying the regulatory regions. We are using sequence changes that arise in rare and common forms of diabetes and hyperinsulinism as a model to gain insights into gene regulation and disease. We study how multiple transcription factors come together to control gene expression, and how changes in regulatory sequences perturb transcription factor binding.
We are using single-molecule footprinting (also known as NOMe-seq, Fiber-seq, SMAC-seq, nanoNOMe, SAMOSA…) to understand how multiple transcription factors bind in pancreas development and function. We use methyltransferases for methylation marks not endogenously present on DNA in human cells (6mA or GpC) to mark accessible base-pairs with methylation. We then use Oxford Nanopore Technologies long-read sequencing to directly read these modifications.
We’re using this and other functional genomic approaches to study transcription factors, cell-cell signalling, and more over pancreatic differentiation and in donor tissue.
We are recruiting multiple positions related to this project:
- Research Technician Job Advert
- A Lab post-doc - pancreatic differentiation and perturbation studies Job Advert
- A computational post-doc - statistical models of single-molecule footprinting data and convolutional neural networks Job Advert
Please contact Nick Owens n.d.l.owens@exeter.ac.uk for more information.
Studying silencing elements
We’re using these tools to understand how transcription factors silence the expression of gene HK1 in pancreatic beta-cells. This builds off our discovery that regulatory variants in a tissue-specific silencing element are a cause of congenital hyperinsulinism published in Nature Genetics.
Primate-specific gene regulation in human pancreas development
We discovered that loss-of-function variants in primate-specific gene ZNF808 neccessary for human pancreas development. ZNF808 is a KRAB zinc finger protein that has co-evolved with the MER11 transposeable element family. The MER11 family has become domesticated, meaning that the MER11 elements act as enhancers and their accesibility is controlled by ZNF808. Our results show that ZNF808 and MER11 have integrated into the human endoderm differentiation network.
This discovery has been recently published in Nature Genetics. We’re now building off this to investigate the mechanisms behind ZNF808 further, we are recruiting a computational post-doc to analyse single-cell transcriptomics and chromatin accessibility to dissect the role of ZNF808 in human development:
- Computational post-doc (fellow or associate): Job Advert
This position is part of a Wellcome Collaborator award, collaboration with Michael Imbeault, Cambridge and Timo Otonkoski & Diego Balboa, Helsinki, together with Elisa De Franco and Andrew Hattersley at Exeter.
Gene Regulatory Defects in disease
We study a range of other gene regulatory defects in disease from congenital heart disease to timing of menopause:
Stankovic S, Shekari S, Huang QQ, Gardner EJ, Owens ND*, Azad A, Hawkes G, Kentistou KA, Beaumont RN, Day FR, Zhao YN. Genetic susceptibility to earlier ovarian ageing increases de novo mutation rate in offspring. medRxiv; 2022
DOISempou E, Kostiuk V, Zhu J, Guerra C, Tyan L, Hwang W, Aguilar EC, Caplan MJ, Zenisek D, Warmflash A, Owens NDL, Khokha MK. Membrane potential drives the exit from pluripotency and the ontogeny of cell fate via calcium and mTOR. Nature Communications. 13,1 6681. 2022
DOI | PubMed | Data and Code